Focusing on the development of industry-leading original detection technologies, and using them to serve public health and life sciences.
Product Center
Assist in the rapid development of small RNA detection applications

-
Plasma/Serum small RNA Extraction Kit
-
small RNA Library Prep Kit For NGS
-
Digital Microfluidic Platform
-
Consumables

Neohalo Biosciences
Hang Zhou Neohalo Biotechnology Co., Ltd. is a wholly-owned subsidiary of New Horizon Health (6606.HK), a listed company on the Hong Kong Stock Exchange. It was established in April 2022 and is located in the China (Zhejiang) Pilot Free Trade Zone.Neohalo Biosciences is a supplier of reagents and instrument platforms focusing on the upstream of nucleic acid research. It is mainly engaged in the development, promotion and application of nanomaterials and biochip products, and thus serves public health and life sciences. The company is especially committed to developing reagents and equipment related to the research of cell-free RNA (cfRNA) that are internationally leading. At present, it has launched a digital microfluidic technology platform based on the principle of dielectric electrowetting, which can be widely used in various scientific research and clinical scenarios such as micro-sample preparation, fluorescence quantitative PCR, digital PCR, NGS and single-cell library construction.Meanwhile, it has also launched a self-developed micro small RNA extraction kit based on the patented technology of Silicon-Magnetic Aggregates (SMAGG) and a micro small RNA library construction kit based on the patented technology of adapter self-ligation product removal.In addition, the company has currently passed the ISO9001 quality management system, which maximally ensures the stable and reliable quality of the developed products and helps to promote the rapid development of the application field of cfRNA detection.
-

R&D Capability
R&D Capability
Neohalo Biosciences focuses on developing industry-leading original detection technologies to serve public health and life sciences
-

Patent system
Patent system
Micro small RNA isolation and purification products based on patented nano magnetic bead technology and micro small RNA library construction products based on patented joint self linking product removal technology.
-

Platform construction
Platform construction
The company also has a 100000 level clean production workshop in Hangzhou, ensuring the stable and reliable quality of the developed products to the greatest extent, and assisting in the rapid development of small RNA detection applications.
INDUSTRY INFORMATION
Assist in the rapid development of small RNA detection applications








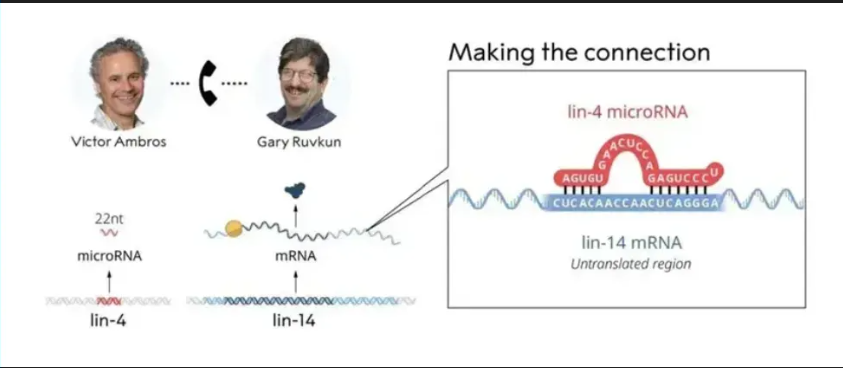

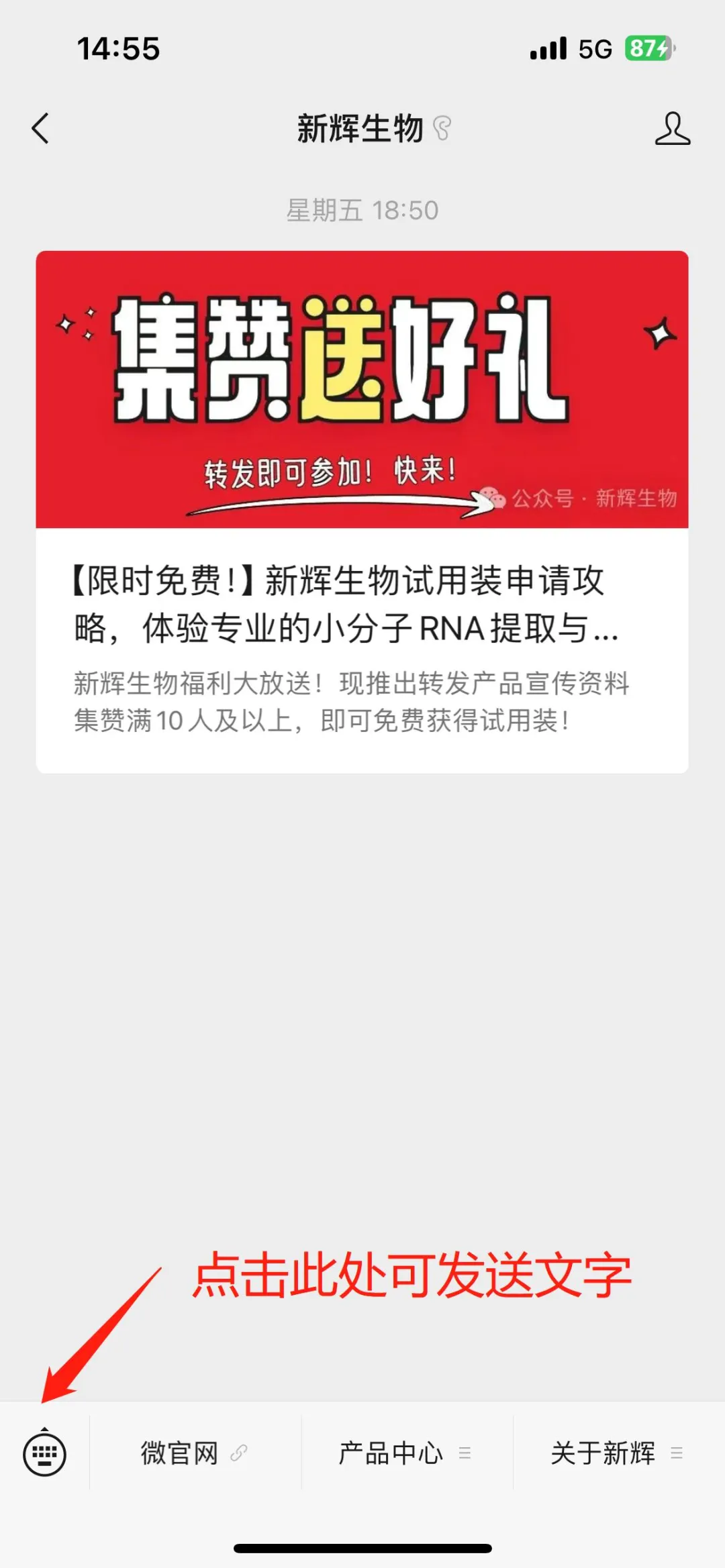











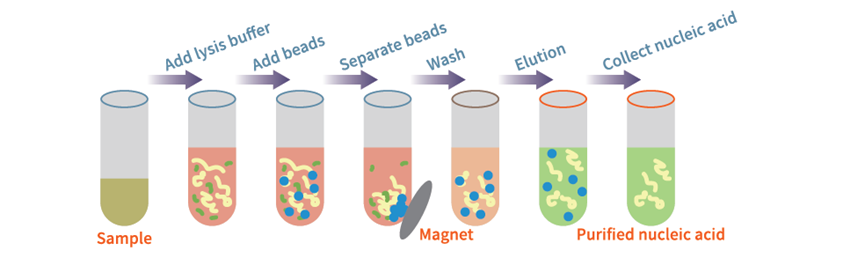
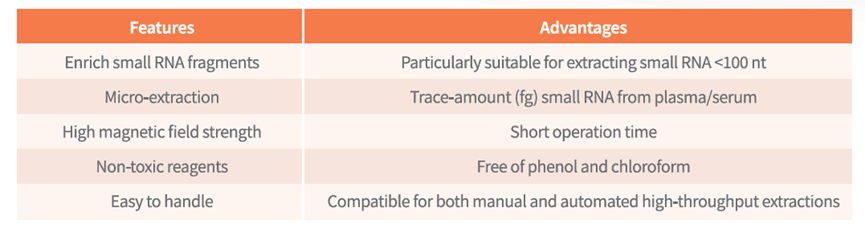
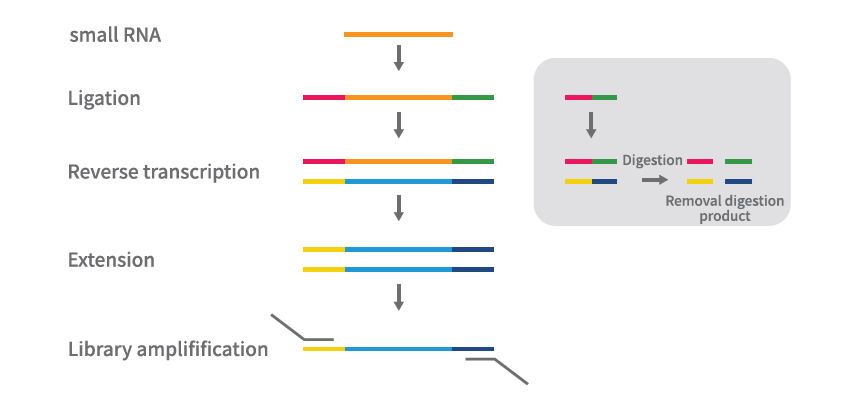
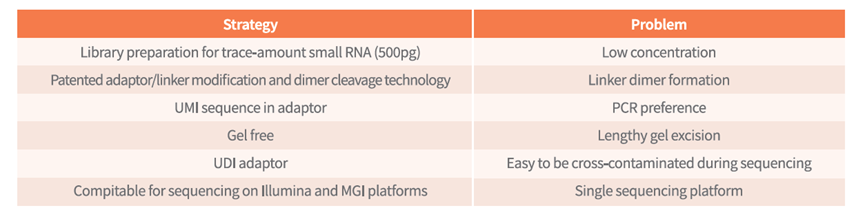




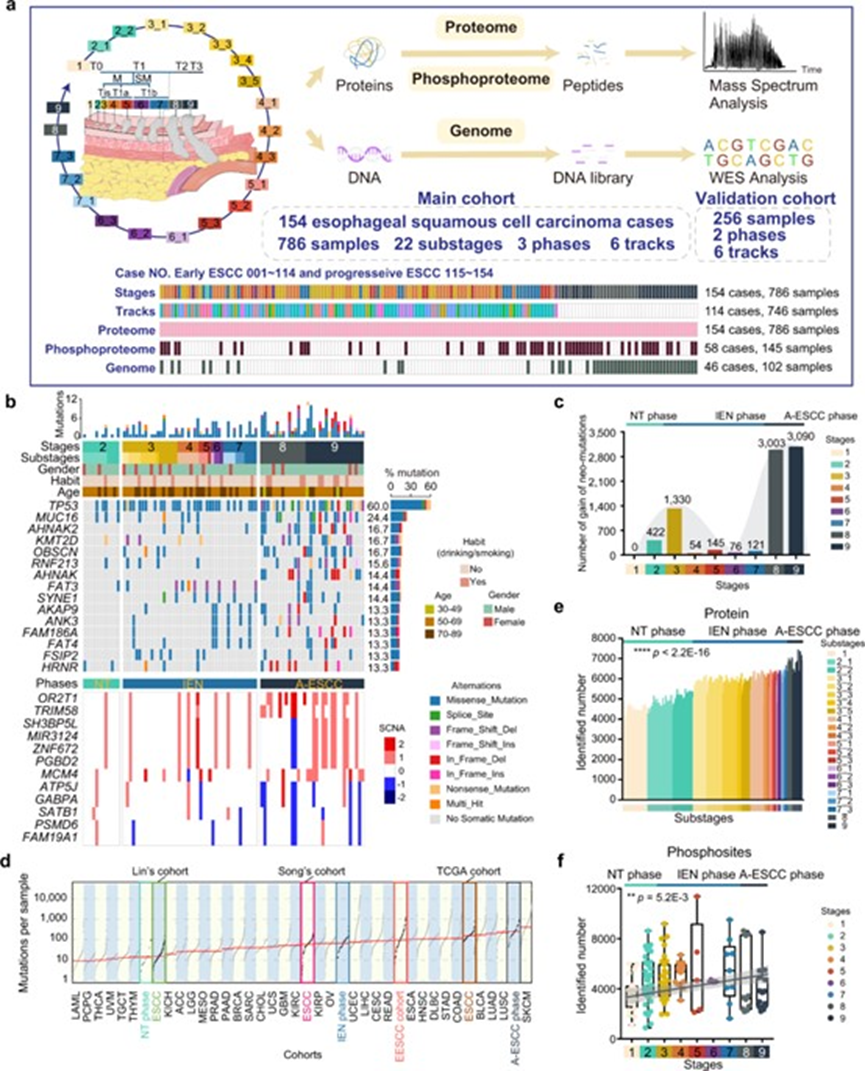

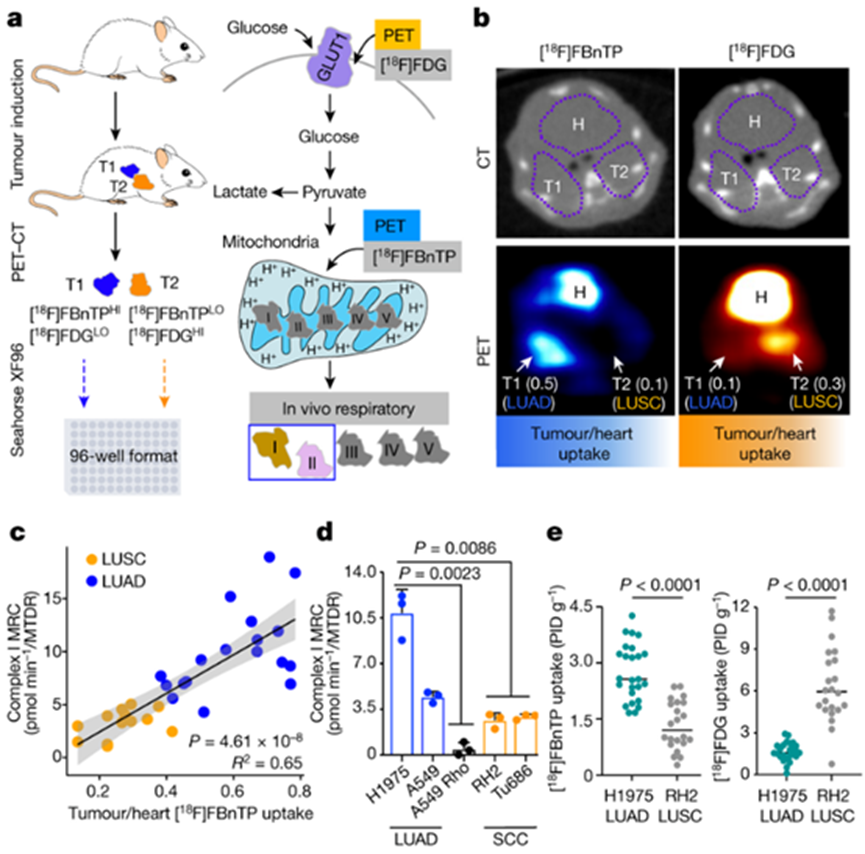 Image source: Nature (2023) DOI: 10.1038/s41586-023-05793-3
Image source: Nature (2023) DOI: 10.1038/s41586-023-05793-3